The 3-R's of data-science:
repeatability, reproducibility & replicability
By Suneeta Mall
Overview
- Industry adaptation
- Importance of 3-Rs
- Peek into Reproducibility crisis
- Define 3-Rs: repeatability, reproducibility & replicability
- Down the memory lane of confused terminology
- Techniques to ensure Repeatability & Reproducibility
- In depth review of few of the promising tools
- Techniques to ensure Replicability
- Few examples
- One last point!
Industry Adaptation
58% of respondents indicated that they were seriously building data science based solutions, with only 14% indicating no involvement just yet.Evolving Data Infrastructure - Ben Lorica and Paco Nathan (O’Reilly, Oct 2018)
Industry Adaptation
As per a survey in UK, 84% of startups focus on Data-science. With 52% of companies preferred to build/use their own models.David Kelnar, MMC Ventures, 2016
Data-Science
Importance of 3-Rs in Data-Science

Know what... why ... & how ...
recreate it ... & solve it.
Importance of 3-R'S .. ctd..
We are continually faced by great opportunities brilliantly disguised as insoluble problems. Lee Iacocca
The opportunities here are building:
Reliable, & robust predictive solution - That can be trusted
IMPORTANCE OF 3-R'S: in research
Non-reproducible single occurrences are of no significance to science.
Popper (The logic of Scientific Discovery)
Yet 70% of researchers have failed to reproduce another scientist's experiments , and > 50% have failed to reproduce their own experiments
- Nature's Survey (2016)
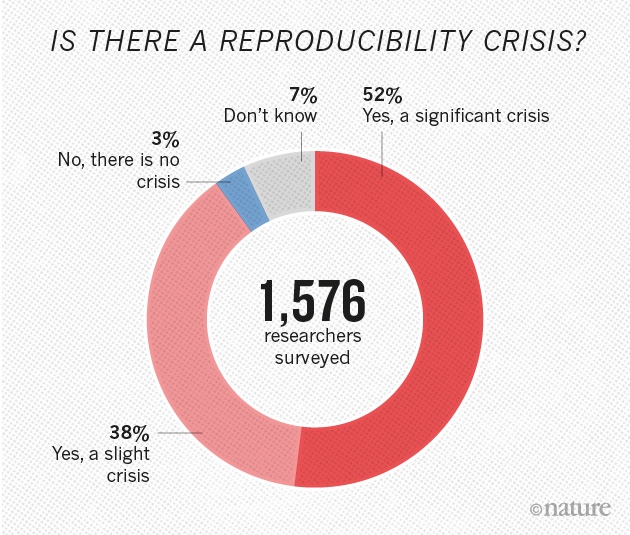
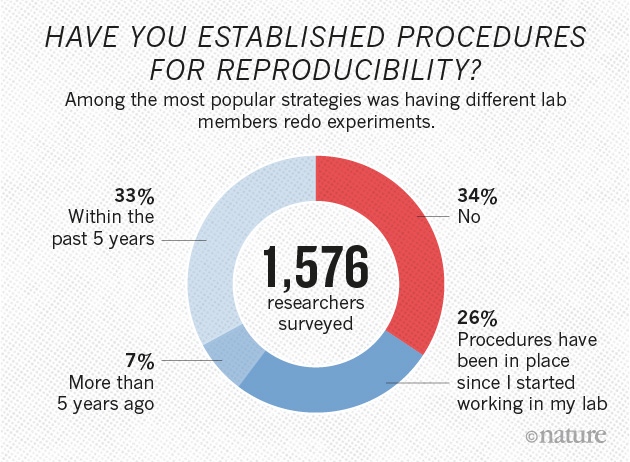
A reproducibility crisis
International Conference on Learning Representations Annual reproducibility challenge (since 2018)lead by Dr. Joelle Pineau, an Associate Professor at McGill University and lead for Facebook’s Artificial Intelligence Research lab (FAIR)
Industry Adaptation is making data-science accessible to people Thus changing our community, and society
We have moral and social obligation to provide confident and reliable answers!
1Repeatability, Reproducibility & Replicability
1.1Repeatability

is the closeness of the agreement between the results of successive attempt of the same experiment/process carried out under the same conditions.
e.g. replay, repeat
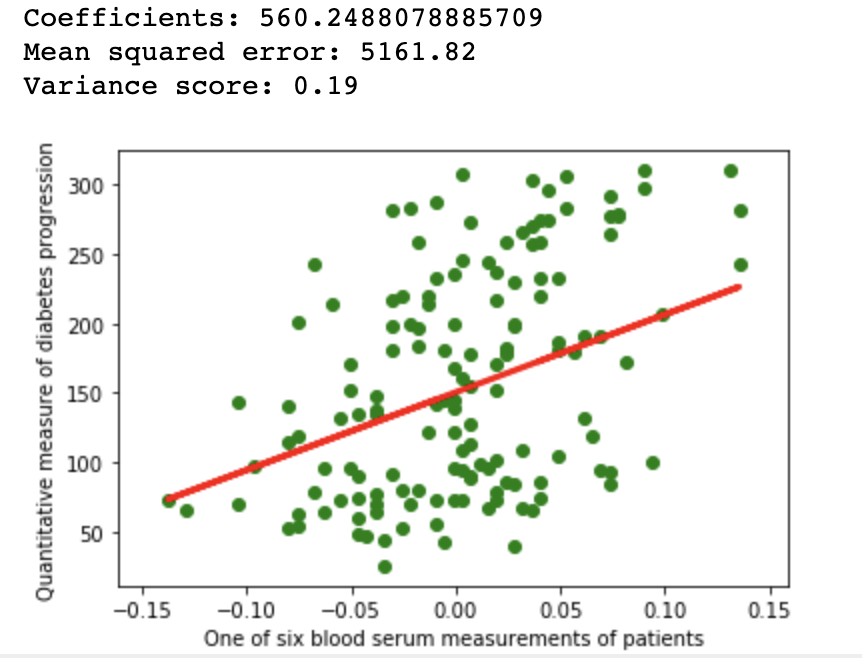
1.1Repeatability
import matplotlib.pyplot as plt
import numpy as np
from sklearn import datasets, linear_model
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.model_selection import train_test_split
diabetes = datasets.load_diabetes()
diabetes_X = diabetes.data[:, np.newaxis, 9]
xtrain, xtest, ytrain, ytest = train_test_split(
diabetes_X, diabetes.target,
test_size=0.33, random_state=None)
regr = linear_model.LinearRegression()
regr.fit(xtrain, ytrain)
diabetes_y_pred = regr.predict(xtest)
# The coefficients
print(f'Coefficients: {regr.coef_[0]}\n'
f'Mean squared error: {mean_squared_error(ytest, diabetes_y_pred):.2f}\n'
f'Variance score: {r2_score(ytest, diabetes_y_pred):.2f}')
# Plot outputs
plt.scatter(xtest, ytest, color='green')
plt.plot(xtest, diabetes_y_pred, color='red', linewidth=3)
plt.ylabel('Quantitative measure of diabetes progression')
plt.xlabel('One of six blood serum measurements of patients')
plt.show()
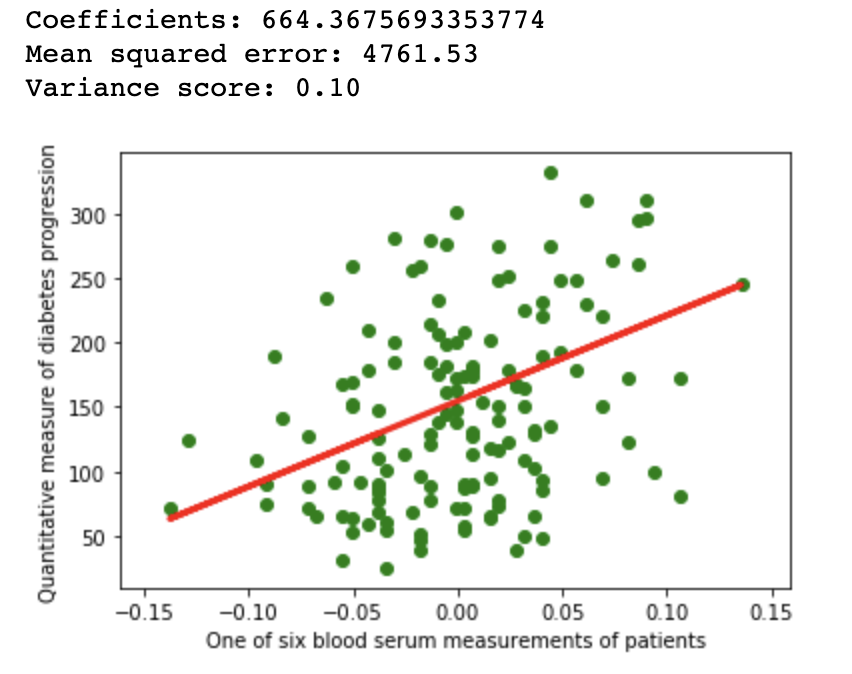
A simple linear regression example on Scikit diabetes dataset
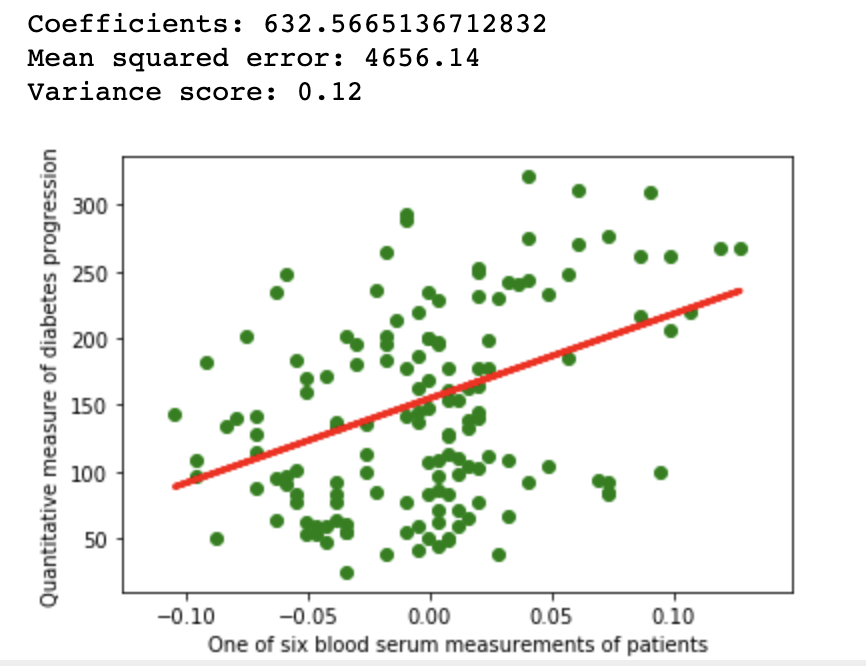
1.1Repeatability
import matplotlib.pyplot as plt
import numpy as np
from sklearn import datasets, linear_model
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.model_selection import train_test_split
diabetes = datasets.load_diabetes()
diabetes_X = diabetes.data[:, np.newaxis, 9]
xtrain, xtest, ytrain, ytest = train_test_split(
diabetes_X, diabetes.target,
test_size=0.33, random_state=32)
regr = linear_model.LinearRegression()
regr.fit(xtrain, ytrain)
diabetes_y_pred = regr.predict(xtest)
# The coefficients
print(f'Coefficients: {regr.coef_[0]}\n'
f'Mean squared error: {mean_squared_error(ytest, diabetes_y_pred):.2f}\n'
f'Variance score: {r2_score(ytest, diabetes_y_pred):.2f}')
# Plot outputs
plt.scatter(xtest, ytest, color='green')
plt.plot(xtest, diabetes_y_pred, color='red', linewidth=3)
plt.ylabel('Quantitative measure of diabetes progression')
plt.xlabel('One of six blood serum measurements of patients')
plt.show()
Linear regression example on Scikit diabetes dataset with fixed seed
1.2Reproducibility
is the closeness of the agreement between the results of successive attempt of the same experiment/process carried out under different conditions.
e.g. repeat*
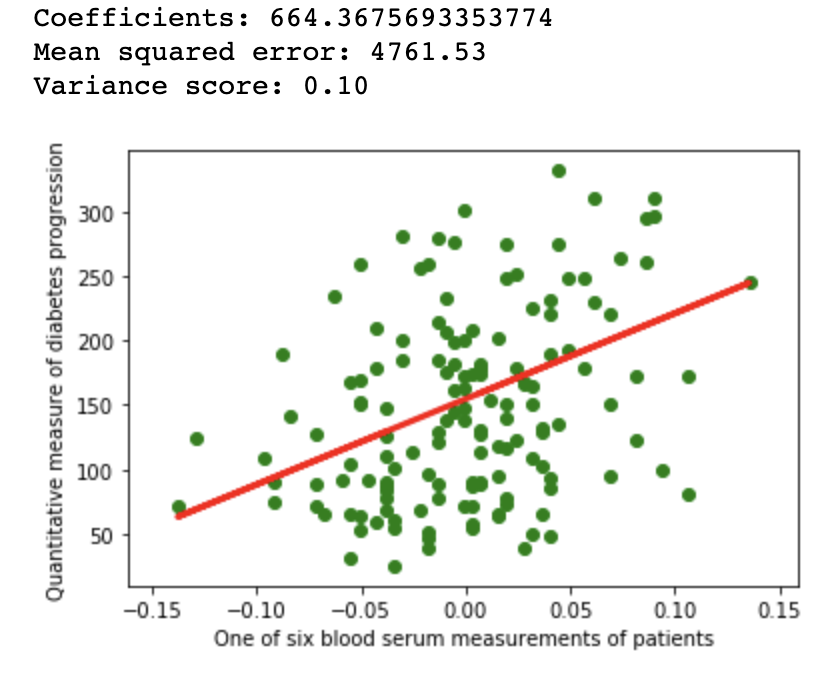
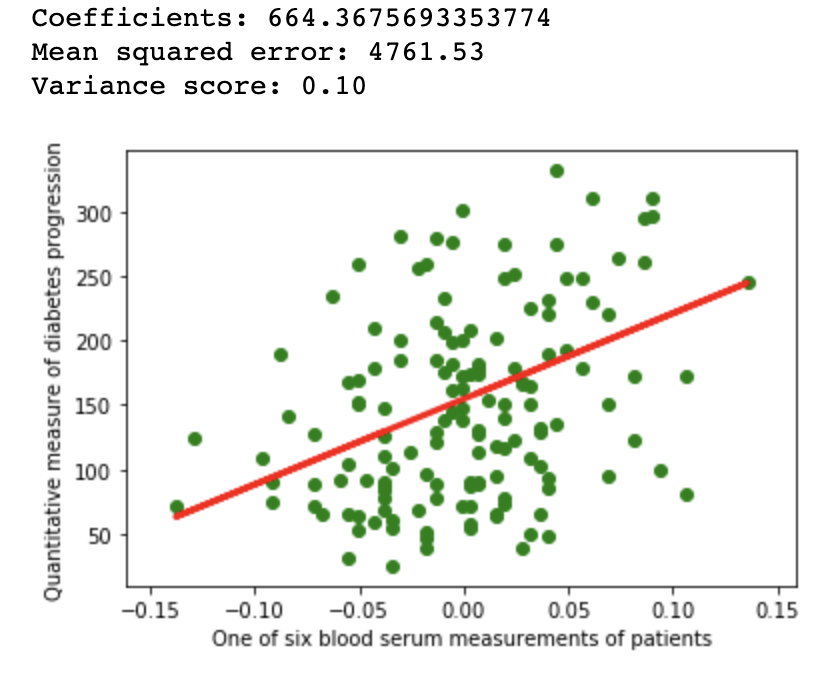
1.2Reproducibility
import matplotlib.pyplot as plt
import numpy as np
from sklearn import datasets, linear_model
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.model_selection import train_test_split
diabetes = datasets.load_diabetes()
diabetes_X = diabetes.data[:, np.newaxis, 9]
xtrain, xtest, ytrain, ytest = train_test_split(
diabetes_X, diabetes.target,
test_size=0.33, random_state=32)
regr = linear_model.LinearRegression(normalize=False)
regr.fit(xtrain, ytrain)
diabetes_y_pred = regr.predict(xtest)
# The coefficients
print(f'Coefficients: {regr.coef_[0]}\n'
f'Mean squared error: {mean_squared_error(ytest, diabetes_y_pred):.2f}\n'
f'Variance score: {r2_score(ytest, diabetes_y_pred):.2f}')
# Plot outputs
plt.scatter(xtest, ytest, color='green')
plt.plot(xtest, diabetes_y_pred, color='red', linewidth=3)
plt.ylabel('Quantitative measure of diabetes progression')
plt.xlabel('One of six blood serum measurements of patients')
plt.show()
Linear regression example on Scikit diabetes dataset with different configuration
1.3Replicability

is the closeness of the agreement between the results of original experiment/process to that of independent experiment/process conducted to simuate/replicate original experiement with at least similar data-set, algorithms, and conditions.
e.g. duplicate, copy
1.3Replicability
History of confused terminology
Plesser HE. Reproducibility vs. Replicability: A Brief History of a Confused Terminology. Front Neuroinform. 2018;11:76. Published 2018 Jan 18. doi:10.3389/fninf.2017.00076
Claerbout, J. F., and Karrenbach, M. (1992). Electronic documents give reproducible research a new meaning. SEG Expanded Abstracts 11, 601–604. doi: 10.1190/1.1822162
Drummond, C. (2009). “Replicability is not reproducibility: nor is it good science,” in Proceedings of the Evaluation Methods for Machine Learning Workshop at the 26th ICML (Montreal, QC). Available online at: http://www.site.uottawa.ca/~ cdrummon/pubs/ICMLws09.pdf
Association for Computing Machinery, 2016, defined:
Patil, P., Peng, R. D., and Leek, J. T. (2016). A statistical definition for reproducibility and replicability. bioRxiv. doi: 10.1101/066803.
Goodman, S. N., Fanelli, D., and Ioannidis, J. P. A. (2016). What does research reproducibility mean? Sci. Transl. Med. 8:341ps12. doi: 10.1126/scitranslmed.aaf5027
Universal Reproducibility

1.1 Ensuring repeatability
- To Err is human: Automate everything
- Avoid adhoc changes
- Reproducible randomness

1.2 Ensuring reproducibility
- Ensuring repeatability
- Maintaining full provenance
- Model managment, tracing
P2 of R2
Principle
Process
P1Automate everything
Introduce DAG pipelines that are Simple, Modular, Pluggable, Scalable, & Reliable

DAG tools for ML related workload
P2Full Provenance

Maintain version control over everything - aka Lineage, Time Travel, Audit
Tools useful for full provenance
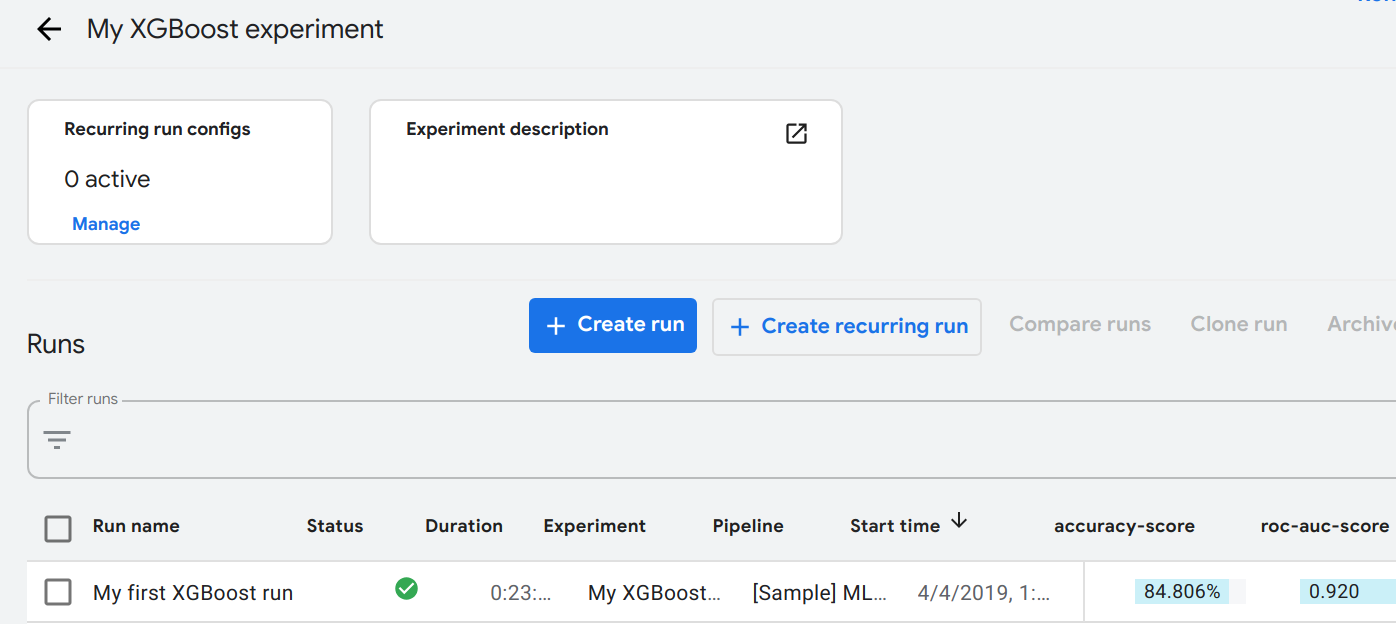
P3Model management - auditing, tracing, serving
Manage gamut of models with: auditing, tracing & serving
Pipeline AI
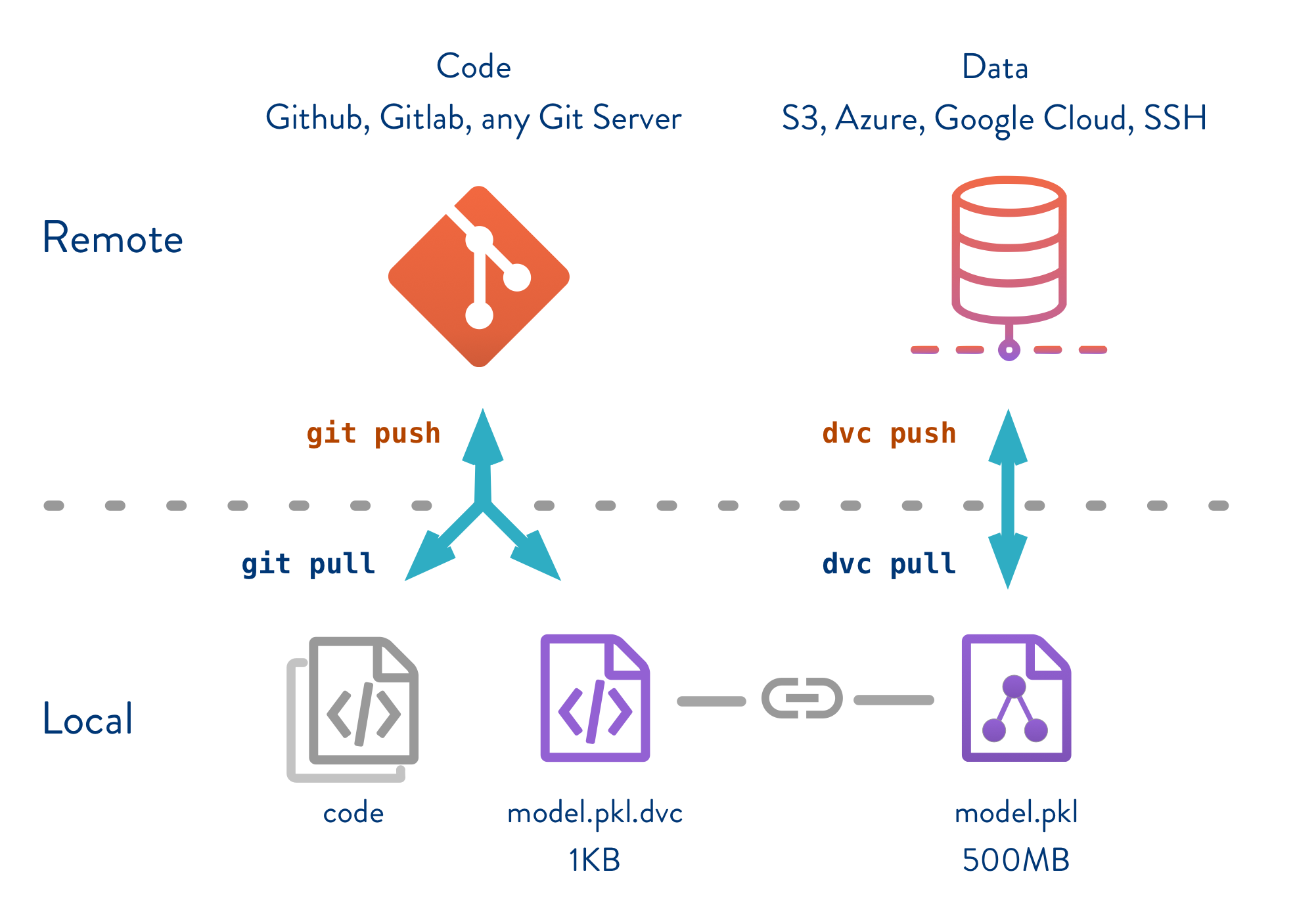
![DVC]() Data Version Control
Data Version Control
Data Version Control
DVC
Pros
- Not very dictative
- Independent tools, integrates easily
- Single source of truth with GitOPS
Cons
- DAG declaration not intutive
- Too much onus on user to integrate with system
- Relatively new - still evolving
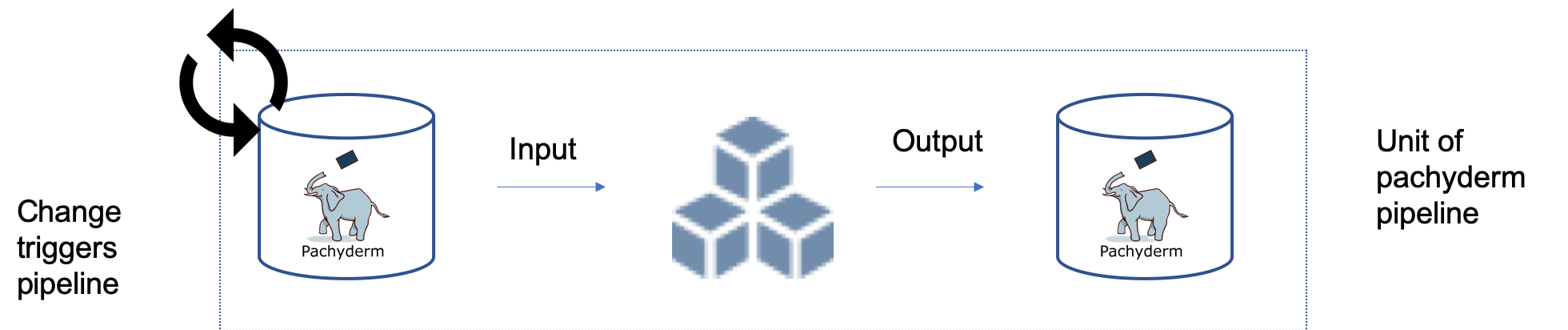
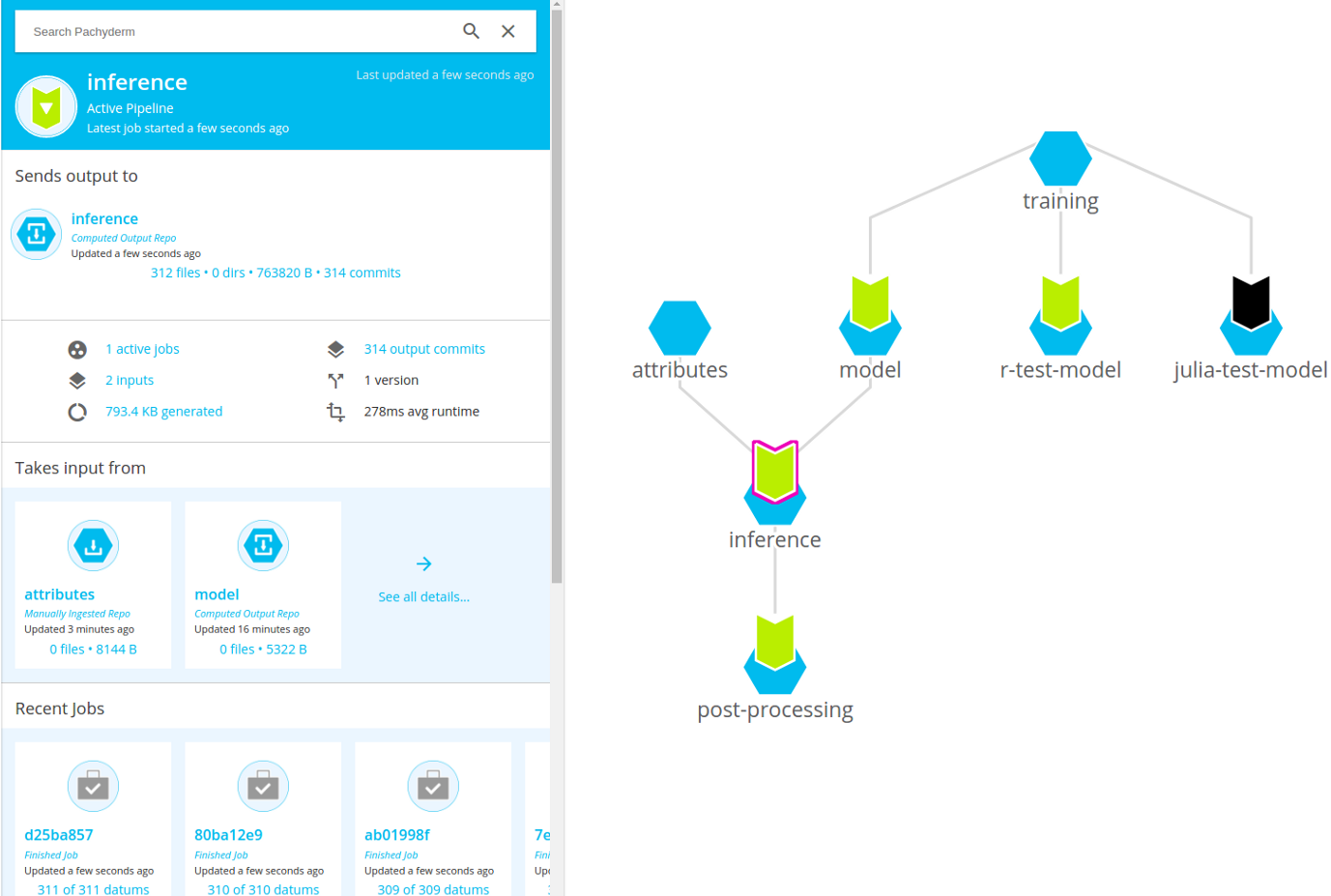
Pachyderm
Pachyderm
{ "input": { "union": [ {
"pfs": {
"glob": "/",
"repo": "training_data",
} }, {
"pfs": {
"glob": "/*.json",
"repo": "parameters",
} } ] },
"pipeline": {
"name": "model_train"
},
"transform": {
"image": "ubuntu",
"cmd": ["/bin/bash"],
"stdin": [
"train_model.py -data
/pfs/training_data
-param /pfs/parameters"
]
}
}
Pipeline Specification
Pachyderm
Pros
- Treats data as first class entity
- Manages versioning automatically
- Simple pipeline constructs
- Scalable & Distributed
- Capability of long running notebooks
Cons
- Is not capable of model serving
- Does not simplify model tracing
- Still rapidly evolving
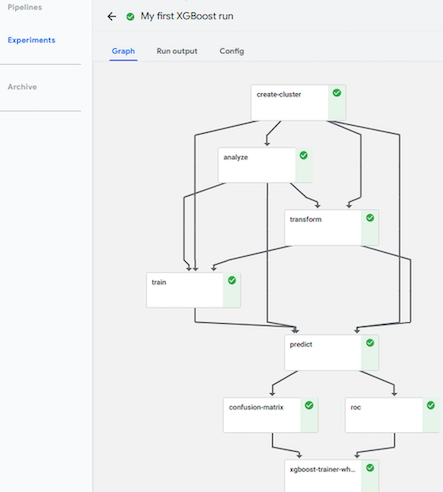
![]() Kubeflow
Kubeflow
Kubeflow
import kfp.dsl as dsl
def PreprocessOp(...):
return dsl.ContainerOp(..)
def TrainOp(...):
return dsl.ContainerOp(...)
@dsl.pipeline(
name='resnet_cifar10_pipeline'
)
def resnet_pipeline(....):
persistent_volume_name = '/'
persistent_volume_path = '/path'
op_dict = {
'preprocess': PreprocessOp(....)
'train': TrainOp(...)
}
for _, op in op_dict.items():
op.add_volume(...)
op.add_volume_mount(...)
if __name__ == '__main__':
import kfp.compiler as compiler
compiler.Compiler().compile(
resnet_pipeline,
__file__ + '.tar.gz')
Pipeline Specification
Kubeflow
Pros
- Leverages kubernetes, scalable, distributed for ML compute
- Nicely integrates with likes of Pachyderm, Katib, ModelDB, Seldon
- GitOPS can be realize with ARGO
Cons
- Does not provide unified provenance
- Requires DSL based pipeline spec
- Installation may feel cumbersome
- Still evolving and integrating with tools
![]() Delta-Lake
Delta-Lake
- Promising offering for Hadoop based workloads
- Leverages spark for data-pipelining
- Manages provenance under time-travel feature
- Model manament/tracing via mlflow
- Model persistence via mleap
Delta Lake
from pyspark.ml import Pipeline
training = spark.createDataFrame(...)
tokenizer = Tokenizer(inputCol="text",
outputCol="words")
hashingTF = HashingTF(...)
lr = LogisticRegression(maxIter=10,
regParam=0.001)
pipeline = Pipeline(stages=
[tokenizer, hashingTF, lr])
model = pipeline.fit(training)
prediction = model.transform(test)
Spark Pipeline Spec
Delta Lake
Only recently launched under Apache 2: 0.1.0 24th April 2019!
- Proprietary version is released a year ago and in use by many organization.
- Very little documentation - so not enough is known
- Claims to have solved the provenance part right!
- Limited to Hadoop/Spark stack
![]() Polyaxon
Polyaxon
- Platform agnostic solution to manage end to end lifecycle of deep leaning workload
- Partly open source with enterprise option
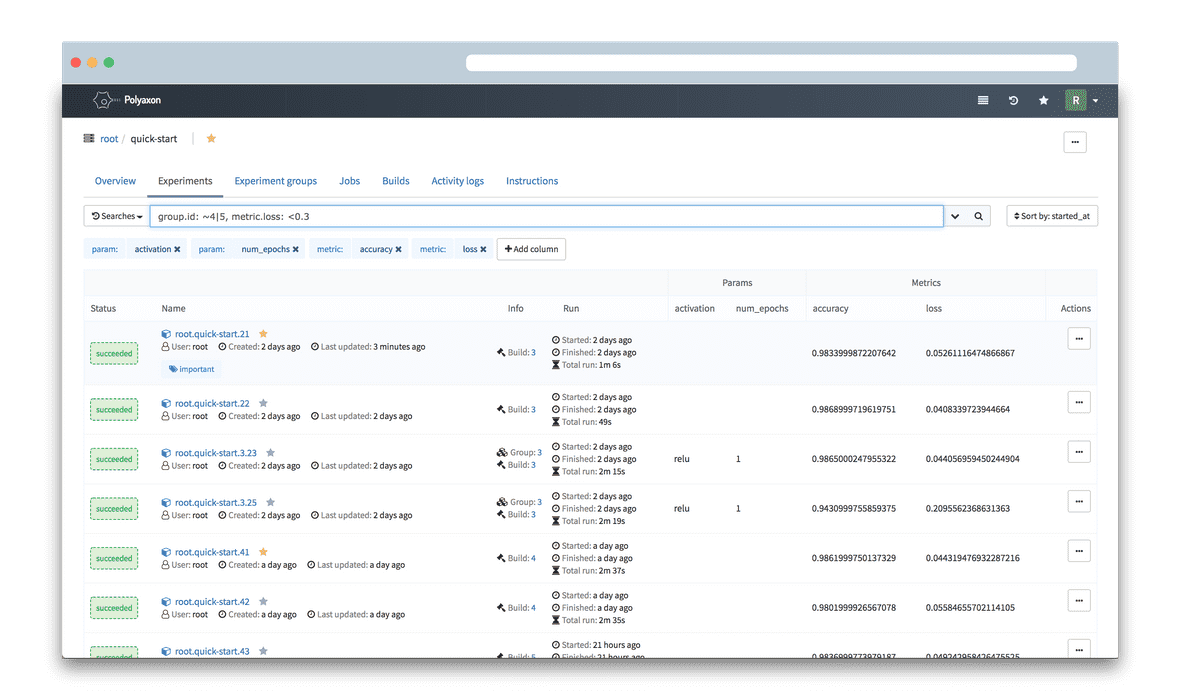
- Provides reproducibility and model tracing
- Integrates with Seldon
- Data provenance is not managed
Polyaxon
1.3Ensuring replicability
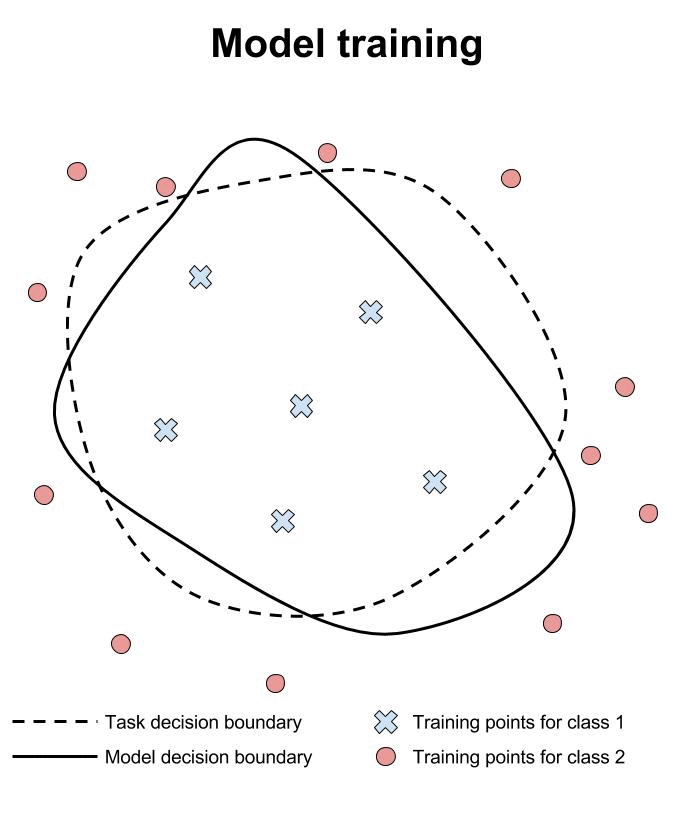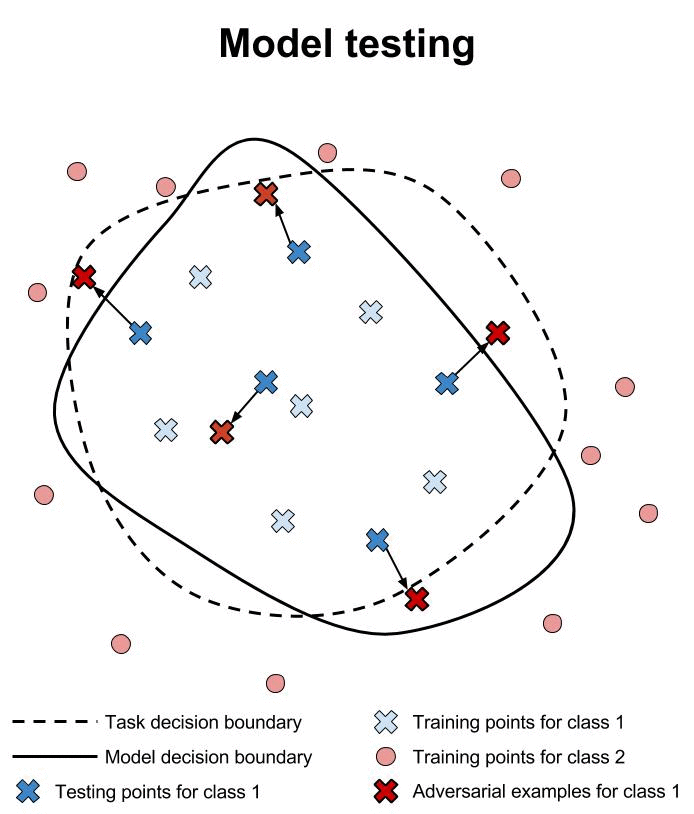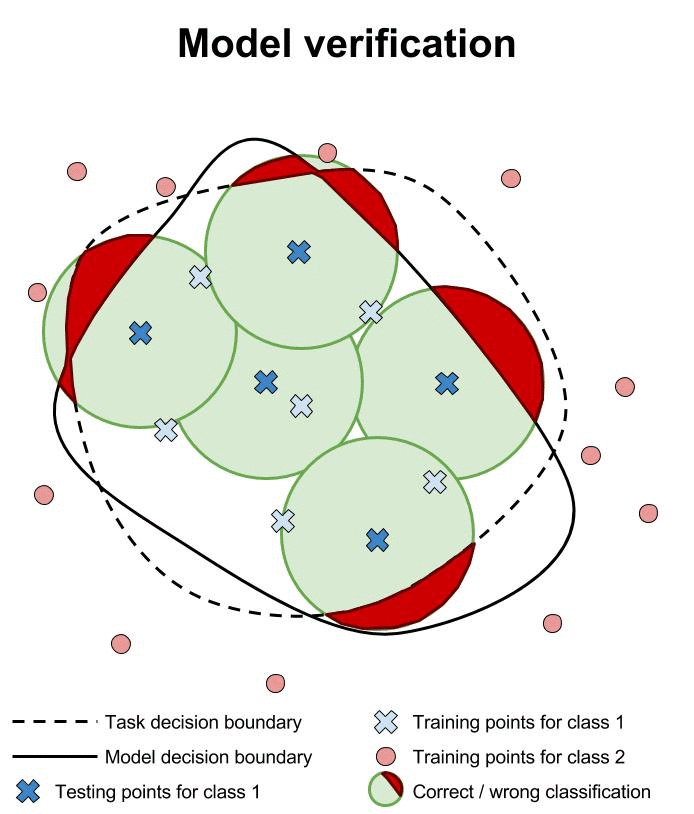
Throwing in 2 more Rs: Reliability and Robustness
1.3Ensuring replicability
Not make confident mistakes (Unrestricted Adversarial Examples - Goodfellow et al.)
Models generalization across architectures and training sets (Explaining and harnessing adversarial examples - Szegedy et al.)
Examples 1
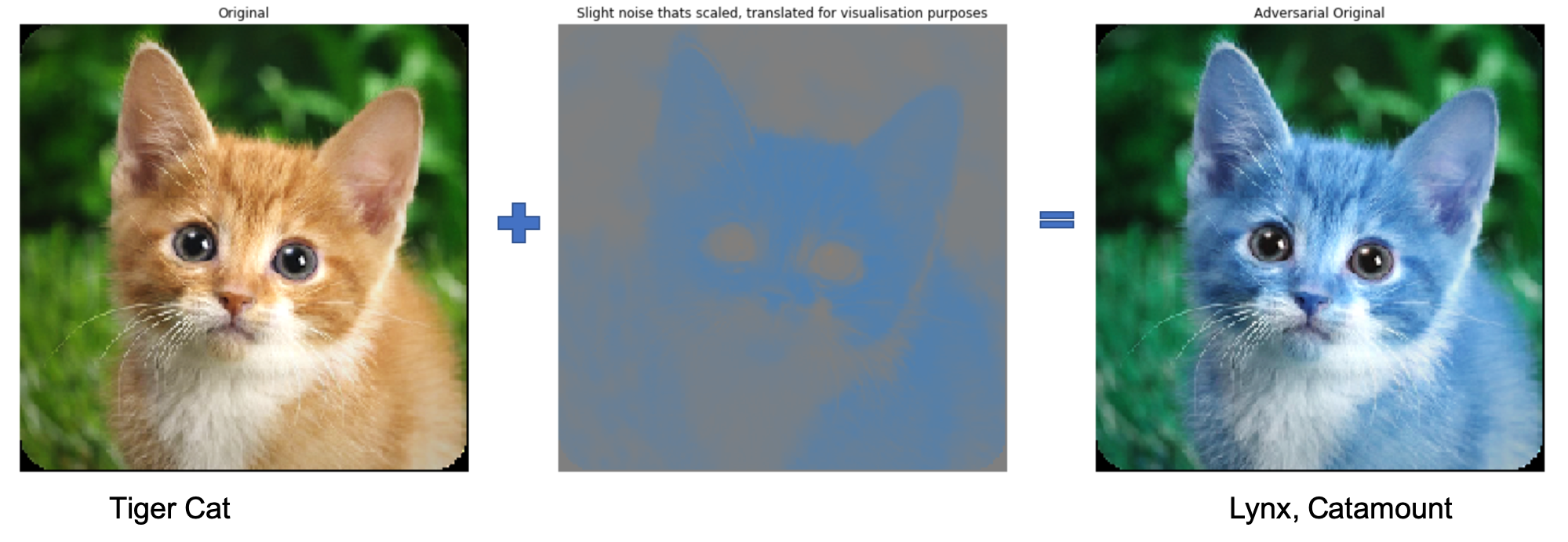
Example 2
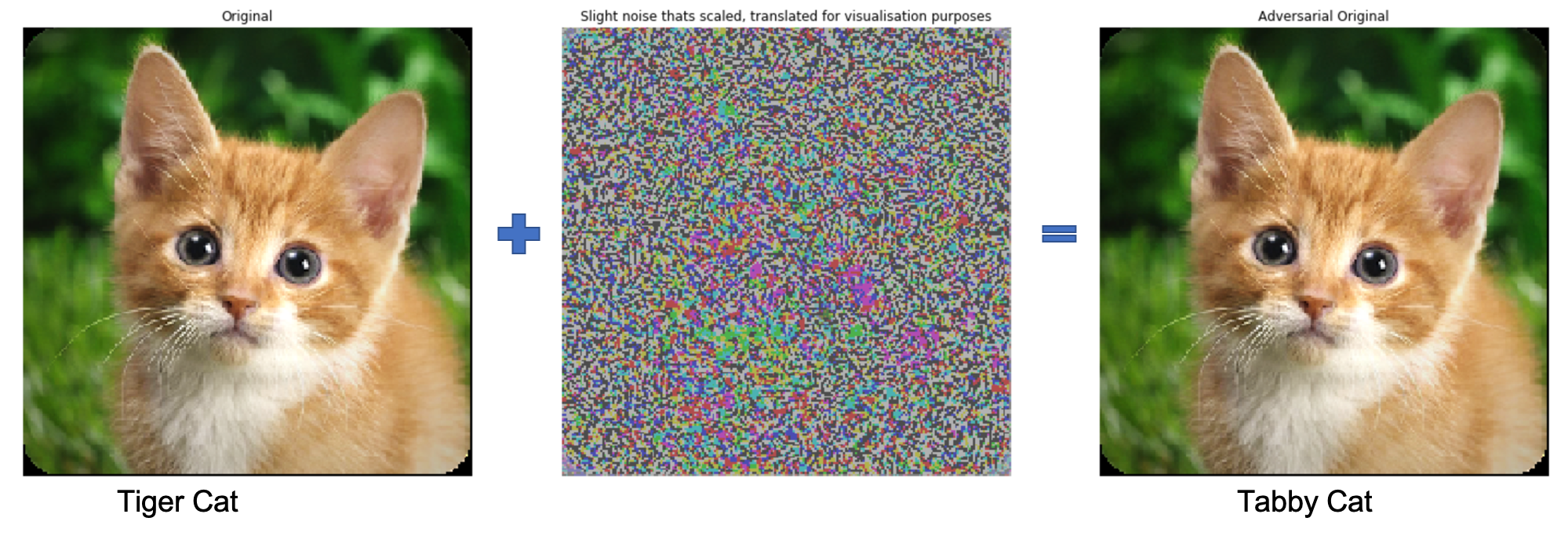
import foolbox
import numpy as np
from foolbox.criteria import Misclassification
from foolbox.models import KerasModel
from foolbox.attacks import FGSM
# model =
fmodel = KerasModel(kmodel, bounds=(0, 255),
preprocessing=(np.array([104, 116, 123]), 1))
attack = FGSM(fmodel, criterion=Misclassification())
# img, label = ,
adversarial = attack(img, label)
Examples generated by foolbox.
Example 3

Lastly
The price of greatness is responsibility.Winston Churchill
Do data-science responsibly!
References & links
| Software | Link | Capability |
|---|---|---|
| DVC | https://iterative.ai/ | DAG, Provenance |
| Pachyderm | http://pachyderm.io | DAG, Provenance |
| Kubeflow | https://github.com/kubeflow/kubeflow | DAG, Model Management |
| Delta Lake | https://delta.io/ | DAG, Provenance, Model Management |
| Pipeline AI | https://github.com/PipelineAI/ | DAG, Model Management |
| Polyaxon | https://github.com/polyaxon/polyaxon | DAG, Model Management |
| Airflow | http://airflow.apache.org/ | DAG, Limited provenance via Atlas DB |
References & links: Part 2
| Software | Link | Capability |
|---|---|---|
| Luigi | https://github.com/spotify/luigi | DAG |
| Nextflow | https://www.nextflow.io/ | DAG |
| Snakemake | https://snakemake.readthedocs.io | DAG |
| Bpipe | http://docs.bpipe.org/ | DAG |
| digdag | https://github.com/treasure-data/digdag/ | DAG |
| Mleap | https://github.com/combust/mleap | Model Persistence |
| Mlflow | https://github.com/mlflow/mlflow/ | Model Management |
| Seldon | https://www.seldon.io/ | Model Management |
| Modeldb | https://github.com/mitdbg/modeldb | Model Management |
| Katib | https://github.com/kubeflow/katib | Hyper parameter tunning |


 Data Version Control
Data Version Control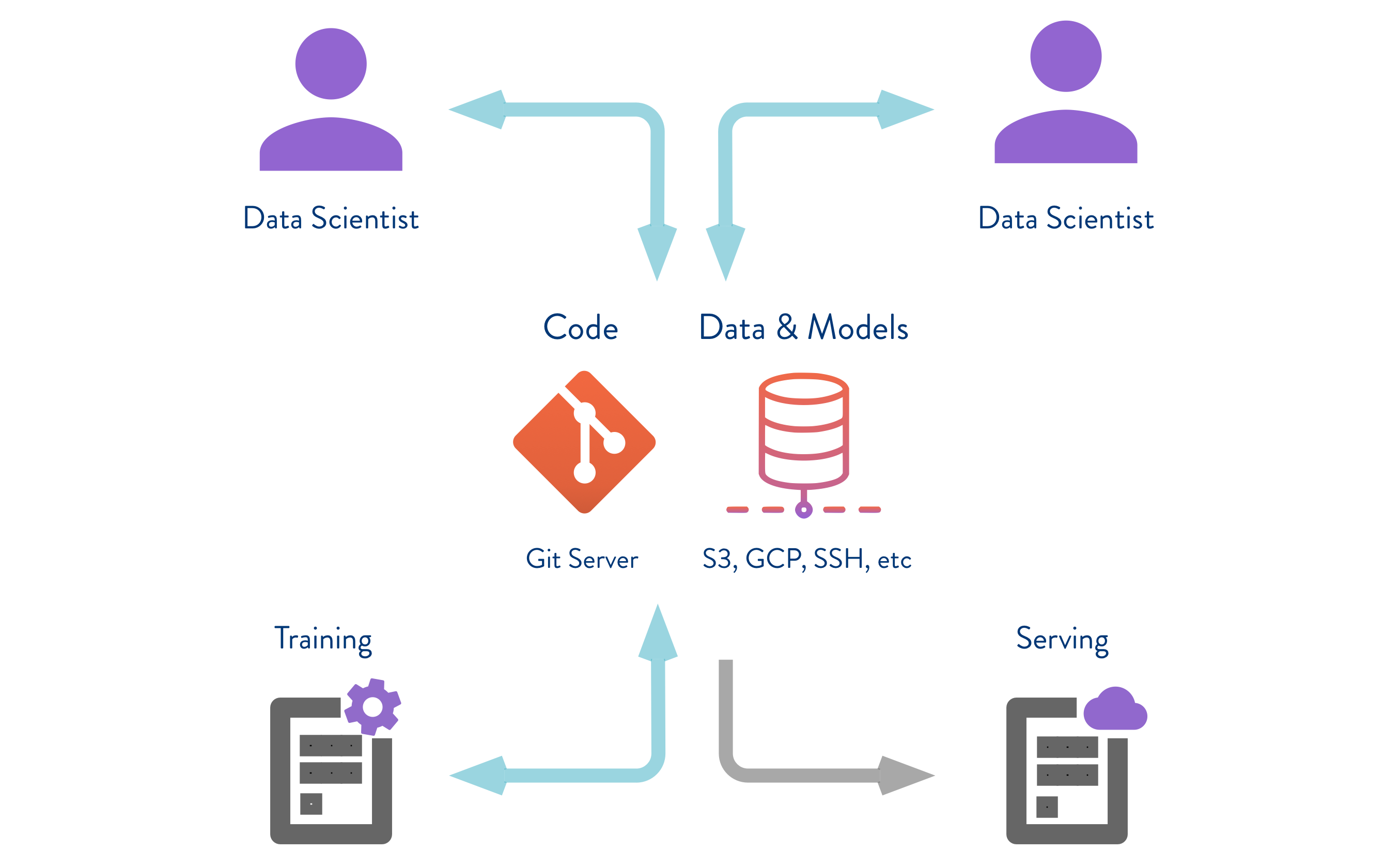
 Delta-Lake
Delta-Lake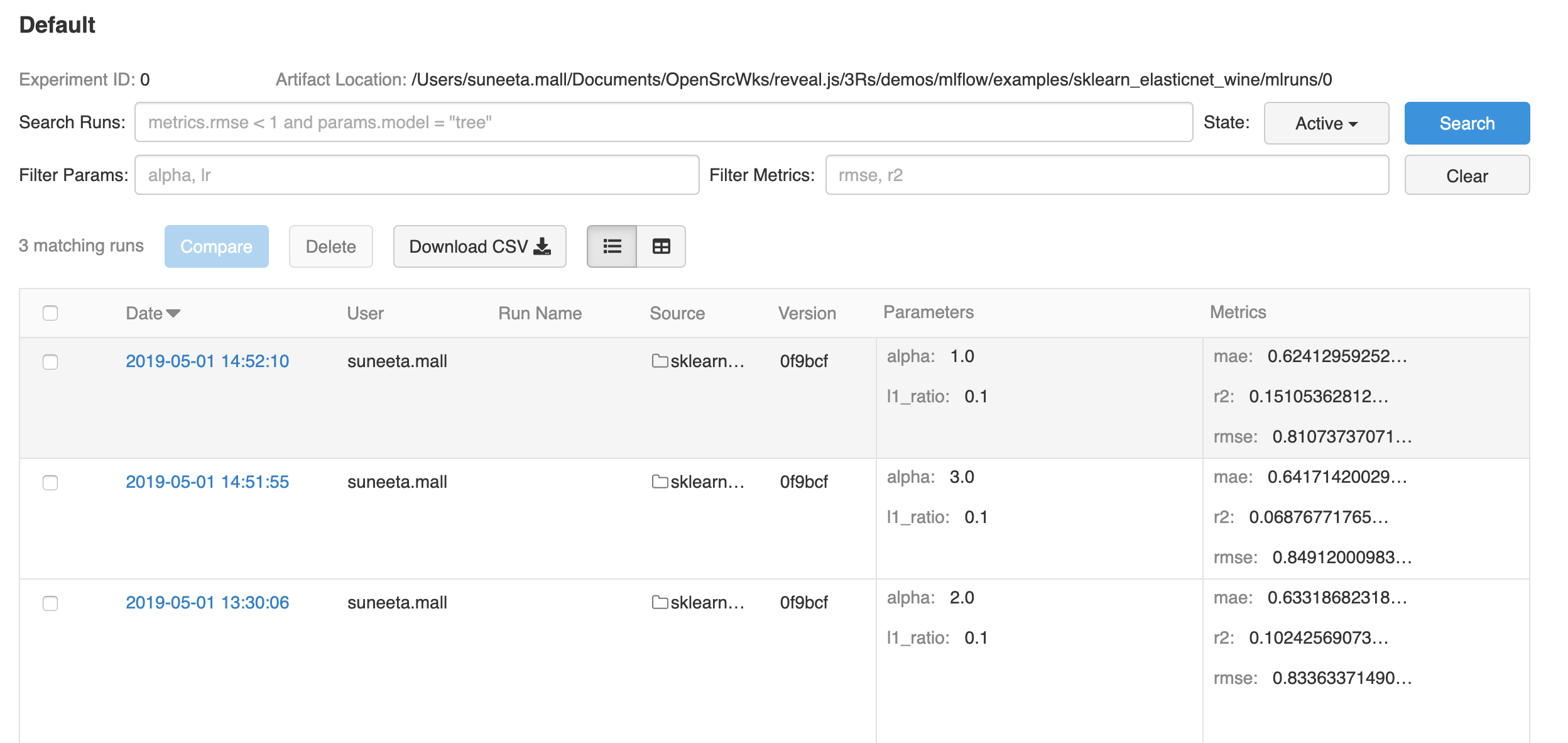
 Polyaxon
Polyaxon